About
I'm a Research Scientist with a background in both Molecular Biology and Bioinformatics. I obtained my PhD degree in Dr. Craig Kaplan's group at University of Pittsburgh (Pitt). During my graduate school study, I dissected Ssl2/TFIIH protein's function in RNA Polymerase II (Pol II) transcription start site (TSS) selection using yeast genetics and NGS methods. After Pitt, I joined Brigham and Women's Hospital as a Research Scientist in Bioinformatics and worked on next-generation sequencing (NGS) data analysis, mainly in single-cell RNA-sequencing data analysis. My expertise involves both dry lab and wet lab skills (moist). Currently, I'm highly interested in using various technologies to solve biological questions that people are most caring about, disease.
Skills
The highlights of my skills are in molecular biology and NGS data analysis. I've also trained by the best Geneticist, my PhD advisor Dr. Craig Kaplan, to learn the power and beauty of the Genetics in solving biological questions. In addition, I love teaching and enjoy the process of interpreting and sharing research with others.
Molecular Biology
- DNA/RNA manipulation
- Molecular cloning
- Genome editing/CRISPR
- In vitro protein expression & purification
- NGS library preparation
- Cell culture
Bioinformatics
- NGS data analysis
- scRNA-seq data analysis
- scATAC-seq data analysis
- Super computing
- AWS
Resume
Education
Doctorate of Science & Biological Sciences
2013 - 2019
University of Pittsburgh, Pittsburgh, PA
Advisor: Craig Kaplan
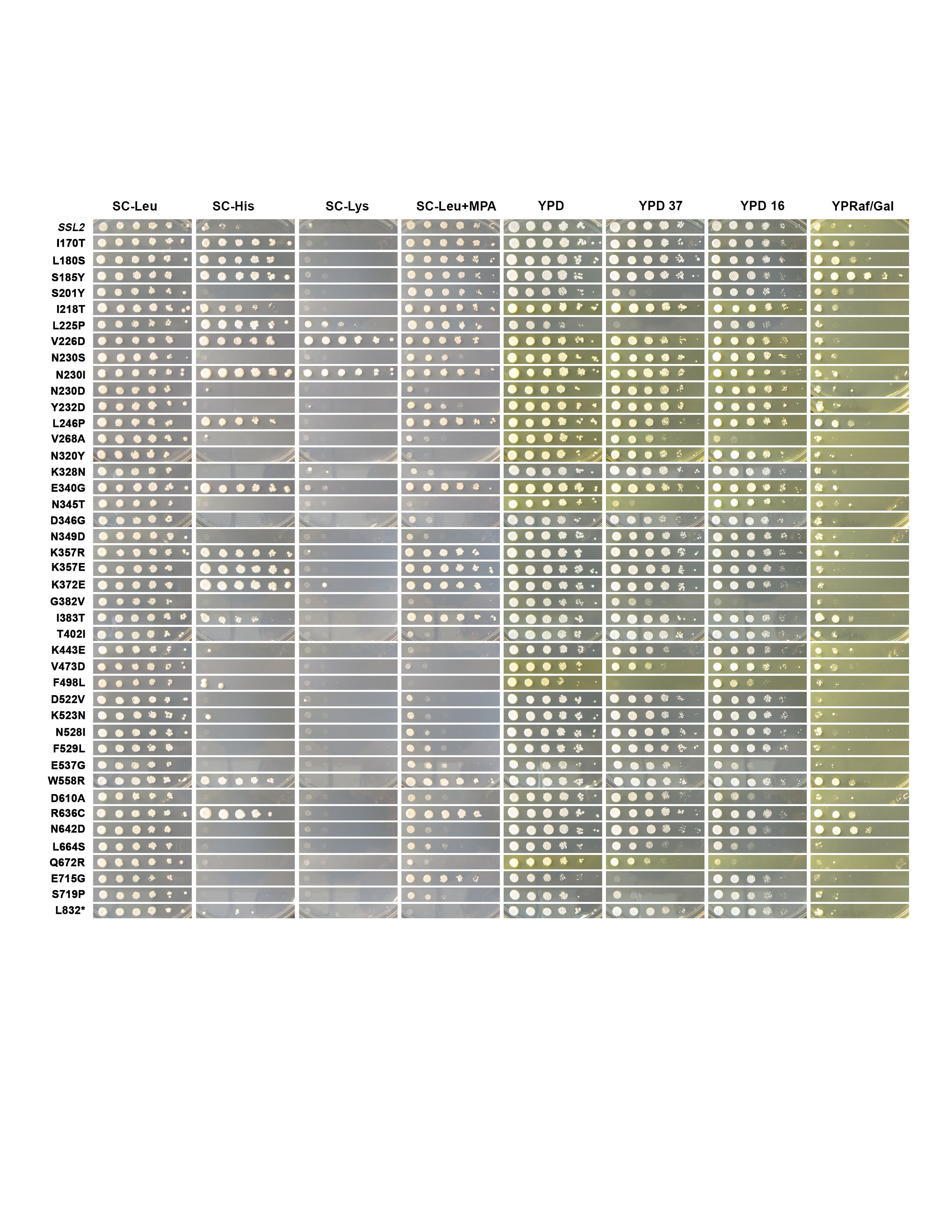
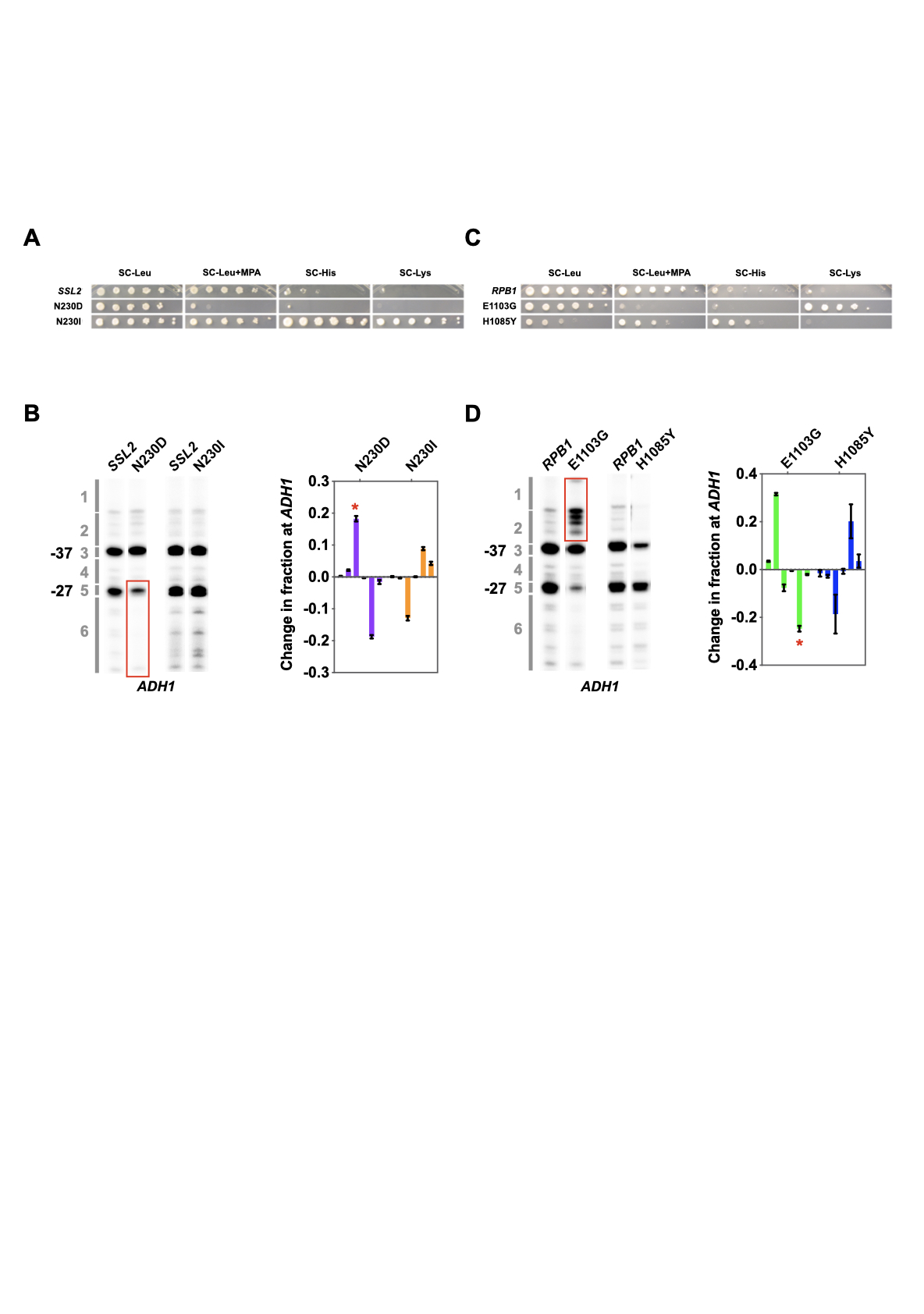
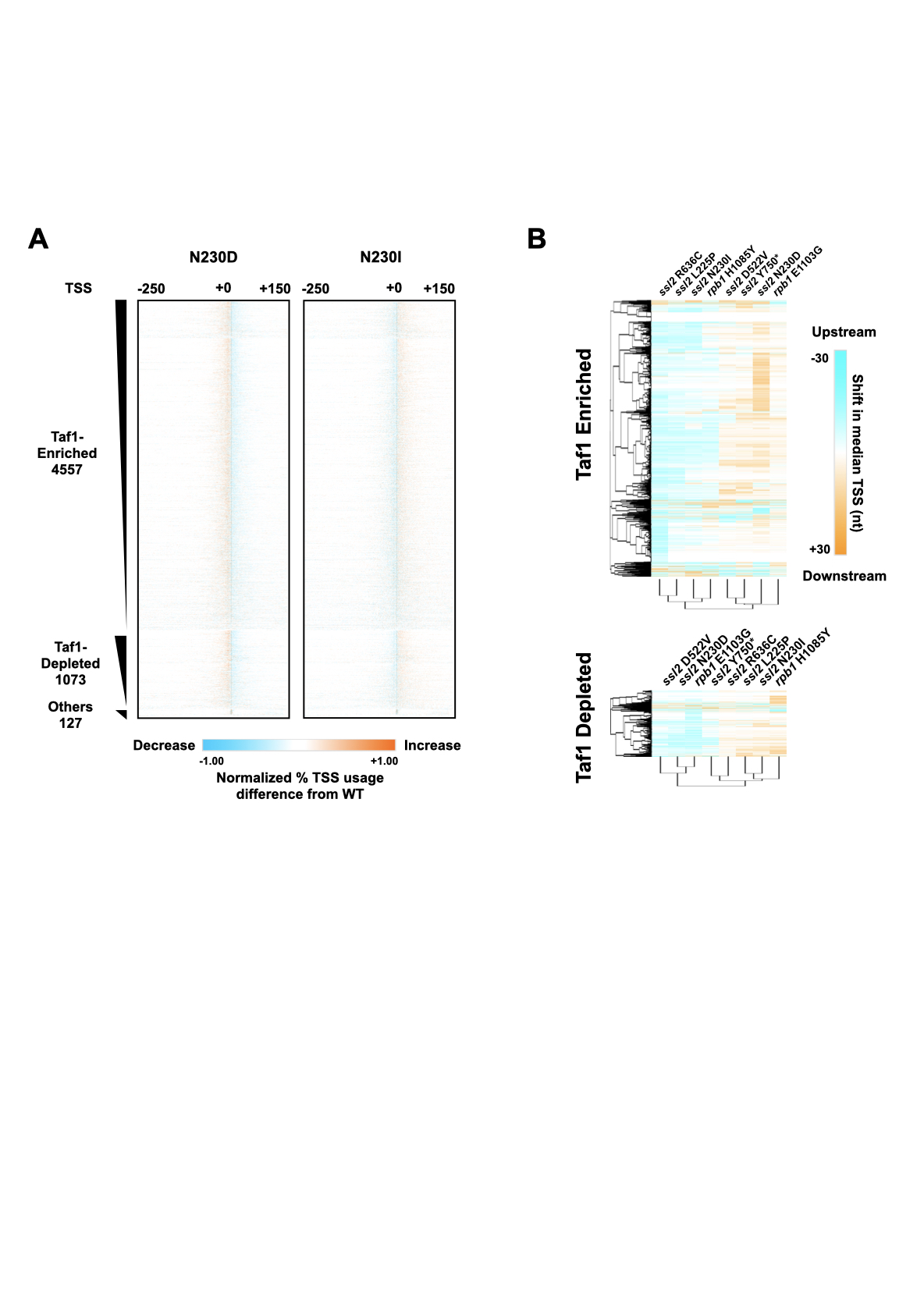
Thesis: Mechanistic insights into the Function of Ssl2/TFIIH in RNA Polymerase II Transcription Start Site Scanning.
Master of Science & Cell Biology
2009 - 2012
Amoy University, Xiamen, China
Advisor: Liang Chen
Thesis: Functional Identification and Expression Analysis in Stress Responses of a NB-ARC Domain Containing Protein OsPDRH9N.
Bachelor of Engineering & Biological Technology
2005 - 2009
Lanzhou University of Technology, Lanzhou, China
Advisor: Xueyan Li
Thesis: Culture condition tests of beer originated Lactobacillus and its exopolysaccharide (EPS) analysis by Thin-layer chromatography (TLC).
Professional Experience
Research Scientist in Medical Oncology
2022 - current
Dana-Farber Cancer Institute, Boston, MA
- Cancer epigenetics.
Research Scientist in Bioinformatics
2021 - 2022
Brigham and Women's Hospital, Boston, MA
- scRNA-seq data analysis for cell type and gene expression investigation.
PhD and Postdoctoral Associate
2013 - 2019
University of Pittsburgh, Pittsburgh, PA
- RNA polymerase II transcription initiation and transcription start site selection.
Research Assistant
2012 - 2013
National Institute of Biological Sciences, Beijing, China
Publications
-
Plasma and CSF biomarkers of aging and cognitive decline in Caribbean vervets. The Journal of the Alzheimer's Association. 2024.
Curran Varma, Eva Luo, Gustaf Bostrom, Praveen Bathini, Daniela Berdnik, Tony Wyss-Coray, Tingting Zhao, Xianjun Dong, Frank Ervin, Amy Beierschmitt, Roberta Palmour, Cynthia Lemere.
-
Ssl2/TFIIH Function in Transcription Start Site
Scanning by RNA Polymerase II in Saccharomyces cerevisiae. eLife. 2021.
Tingting Zhao, Irina O. Vvedenskaya, William K.M Lai, Shrabani Basu, B. Franklin Pugh, Bryce E. Nickels and Craig D. Kaplan.
-
Structure of TFIIH/Rad4-Rad23-Rad33 in damaged DNA
opening in Nucleotide Excision Repair. Nature Communications. 2021.
Trevor van Eeuwen, Yoonjung Shim, Hee Jong Kim, Tingting Zhao, Shrabani Basu, Benjamin A. Garcia, Craig D. Kaplan, Jung-Hyun Min & Kenji Murakami.
-
Universal promoter scanning by Pol II during
transcription initiation in Saccharomyces cerevisiae. Genome Biology, 2020.
Chenxi Qiu , Huiyan Jin , Irina Vvedenskaya , Jordi Abante Llenas, Tingting Zhao , Indranil Malik, Alex M. Visbisky , Scott L. Schwartz , Ping Cui , Pavel Čabart , Kang Hoo Han , William K. M. Lai, Richard P. Metz , Charles D. Johnson , Sing-Hoi Sze, B. Franklin Pugh, Bryce E. Nickels and Craig D. Kaplan.
-
Two novel NAC transcription factors regulate gene
expression and flowering time by associating with the histone demethylase JMJ14. Nucleic Acids
Research. 2015.
Yong-Qiang Ning, Ze-Yang Ma, Huan-Wei Huang, Huixian Mo, Ting-ting Zhao, Lin Li, Tao Cai, She Chen, Ligeng Ma and Xin-Jian He.
Projects conducted at DFCI
-
The effect of BCOR rapid loss on RNA polymerase II transcription and chromatin architecture in leukemia cells. 06/13/22 - 09/06/2024
Using CRISPR, Co-IP, ChIP-qPCR and ChIP-seq technologies to study the mechanisms of BCOR protein in maintaining functional hematopoiesis and how defects of BCOR cause diseases such as myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML). These findings supported a lab grant application in May 2024.
-
High-throughput profiling of RUN1 and GATA2 point mutations and their transcriptional effects in leukemia cells using synthetic variant library approach. 06/13/22 - 09/06/2024
In collaboration with Twist Biosciences, I worked on generating and analyzing leukemia cell lines with patient-derived GATA2 mutants to study their effects on gene expression.
-
Bulk RNA-seq pipeline development. 01/04/24 - 05/05/24
The RNA-seq pipeline developed was used for the BCOR project data analysis, as well as for data analysis in Fred Tsai's and Rahul Vedula's projects.
-
ChIP-seq data analysis. 06/13/24 - 09/06/24
Using published ChIP-seq datasets, I analyzed Pol II occupancy at PRC1.1 target promoters, confirming reduced Pol II binding at BCOR-bound, repressed promoters.
-
Patient-derived RUNX1 mutant characterization. 12/12/22 - 02/17/24
Mentoring Koissi Savi for molecular biology experiments to characterize patient derived RUNX1 single mutant in leukemia cell lines using molecular cloning and genome editing approaches.
Projects conducted at BWH
-
scRNA-seq data analysis to investigate Rattus
norvegicus cell type and gene expression. 02/01/22 - 05/18/2022
scRNA-seq project collaborated with Susanne Wolf at Charite University, Berlin.
-
scRNA-seq data analysis to investigate
Drosophila brain cell type and gene expression. 10/15/21 - 05/12/2022
scRNA-seq project collaborated with Mel Feany at Department of Pathology, Harvard Medical school.
-
scRNA-seq workshop. 10/04/21 - 10/05/21
Teaching assistant for scRNA-seq workshop hold for Harvard community. Project led by Martin Hemberg at Evergrande Center for Immunologic Diseases, Harvard Medical school.
-
Searching for potential Alzheimer's disease
biomarkers through vervet biomarker and behavior analysis. 09/13/21 - 12/31/21
Vervet project collaborated with Cindy Lemere at Brigham and Women's Hospital, Harvard Medical School.
-
Spatial transcriptomic data analysis:
Multiplexed error-robust FISH (MERFISH). 05/12/21 - 09/17/21
MERIFISH project led by Jeffrey Moffitt at Boston Children's Hospital, Harvard Medical School.
-
snRNA-seq data analysis to investigate mouse
pancreas cell type and gene expression. 03/18/21 - 09/10/21
snRNA-seq project collaborated with Sahar Nissim at Boston Children's Hospital, Harvard Medical school.
Conferences
-
Boston Computational Biology and
Bioinformatics (BCBB), Online Lightning Talks, SPEAKER, Aug 12, 2020.
Tingting Zhao, Craig D. Kaplan. Single Mutations in Ssl2 Alter RNA Polymerase II Transcription Start Site Usage Genome-wide.
-
Cold Spring Harbor Laboratory Meetings,
Mechanisms of Eukaryotic Transcription, POSTER, Aug 27 - Aug 30, 2019.
Tingting Zhao, Craig D. Kaplan. Competition and Cooperation Between Activities of Ssl2/TFIIH and RNA Polymerase II in Transcription Start Site Scanning.
-
Chromatin and Epigenetic Regulation of
Transcription, 38th Summer Symposium in Molecular Biology, POSTER, Jul 30 - Aug2, 2018.
Tingting Zhao, Craig D. Kaplan. Mechanistic Insights into the Function of Ssl2/TFIIH in RNA polymerase II Transcription Start Site Scanning.
-
TAMU Student Research Week, SPEAKER, Mar
22, 2017.
Tingting Zhao, Craig D. Kaplan. Function of Ssl2 in RNA Polymerase II Transcription Start Site Scanning.
-
TAMU BioBio Yeast Meeting, SPEAKER, Feb 21,
2017.
Tingting Zhao, Craig D. Kaplan. Function of Ssl2 in RNA Polymerase II Transcription Start Site Scanning.
-
The Allied Genetics Conference 2016 (TAGC),
POSTER, Jul 13 - 17, 2016.
Tingting Zhao, Craig D. Kaplan. Screening of ssl2 alleles with transcriptional defects.
Teaching & Mentoring
-
Gene Team Summer Program, University of
Pittsburgh. July, 2020.
Students' presentations are here: https://www.youtube.com/watch?v=Rfqfx_yz3Xw&list=PLDiSwjR0jbGiSWchh2O0F_r0pRbIunKHY&index=3&t=0s.
-
Training undergraduate student Luis
Lopez Gonzalez for general lab techniques such as PCR, molecular cloning and basic
Drosophila genetics. Aug 2018 - Dec 2018.
Luis is now back to UK.
-
Guiding 4 graduate students to perform
lab research on screening of tfb3 alleles, encoding mutant forms of the general
transcription factor Tfb3, and test their effects on gene expression during their lab
rotations. Sep 2015 - May, 2016.
This research is not published yet.
-
Recitation of Comprehensive
Biochemistry BICH 410 and BICH 411, for ~140 undergraduate students. Fall 2014- Spring
2015.
Students’ evaluations are here: https://docs.google.com/document/d/1npis23uzOvHLRmoSj8Aj7EPn2r6Kt5B6pPC9X1e6BIo/edit.
Growing random thoughts
-
2022, year of changing.
Learning new skills is always good, however, chasing on new technologies is not wise. Mastering how to use diverse technologies into a project for solving a practical question is the goal of my next career stage.
-
2019, year of graduation.
Learning is an endless process, keep learning but also thinking about finding a "place" to land what I have learned.